|
Click to listen to this article
|
Using DNA Evidence to Address Cases of Mistaken Identity of Bacteria Causing Onion Diseases
By Brian Kvitko, University of Georgia; Christy Hoepting, Cornell Cooperative Extension; and Lindsey du Toit, Washington State University
We have shared several reports in Onion World over the past few years on the “Stop the Rot” onion bacterial project. The project has two main areas of focus on bacterial diseases that affect onion production across the U.S.: 1) understanding the diversity and prevalence of bacteria that cause onion diseases, and 2) determining how to manage these bacterial pathogens more effectively.
As part of the first objective, we are taking a close look at the genetics (DNA) of the numerous bacteria we have found in onion crops surveyed around the U.S. over the past three years. You might ask “So what? Why is this important in order to manage onion bacterial diseases?”

Distinguishing the Good From the Bad
Genetics of onion bacteria are important to distinguish the good from the bad. Many species of bacteria can cause diseases of onion. Even more bacteria can make a “happy home” inside a rotting onion once another pathogen starts to break down onion tissue. This can make it extremely difficult to get an accurate diagnosis of which bacterium is the primary culprit for causing an onion to rot and which ones are “along for the ride.”
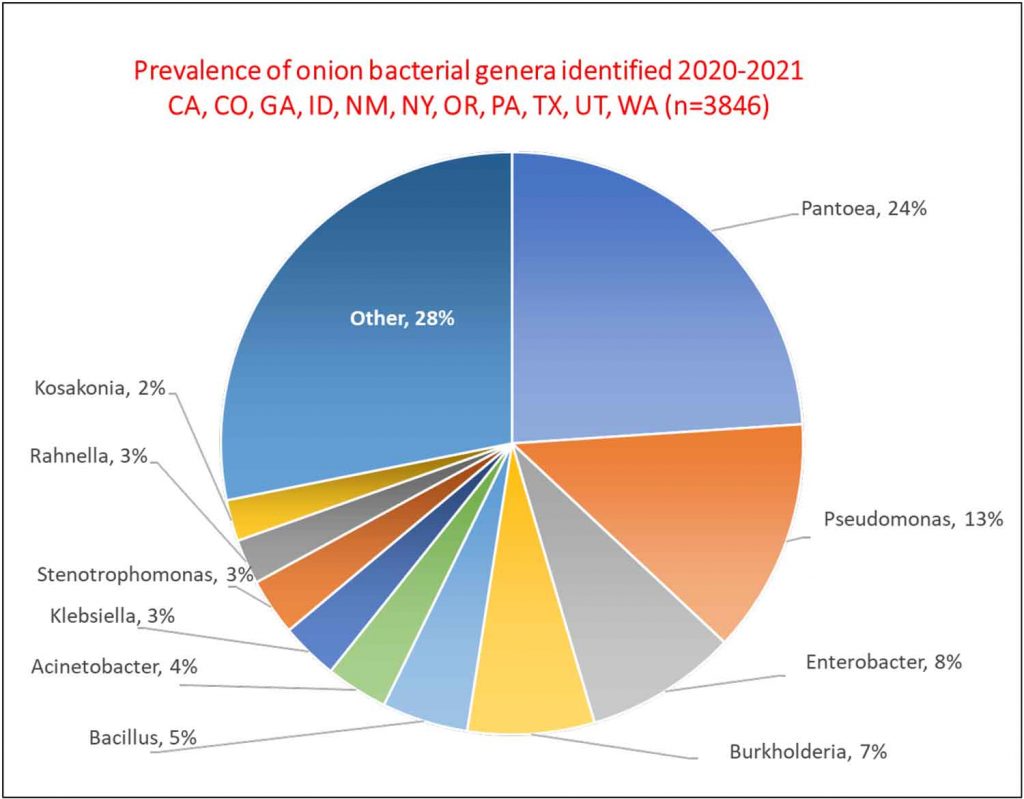
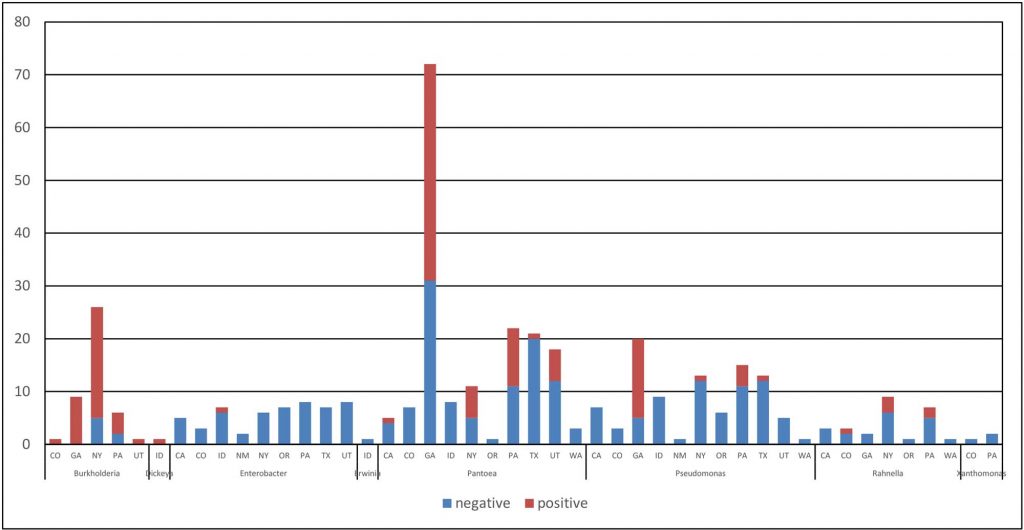
In three years of onion surveys for the Stop the Rot project, we have isolated more than 5,000 bacterial strains that belong to more than 110 different genera. A majority of these bacterial isolates do not cause onion diseases.
Sometimes, diagnosing the species of the bacterium is not enough. A prime example is the center rot pathogen, Pantoea ananatis. We routinely recover isolates of P. ananatis that are aggressive pathogens of onion, but we also routinely recover isolates of P. ananatis that are harmless (not pathogenic) as they are not able to cause disease symptoms on onion bulbs or plants. Sometimes, pathogenic and non-pathogenic strains of P. ananatis can be isolated from the same onion plant or bulb. This is similar for several other bacterial species with strains that can cause onion diseases and strains that are not pathogens of onion. This kind of situation can lead to mistaken identity and misdiagnosis of the primary pathogen; a strain that is “along for the ride” may get “accused of committing the crime.”
The variation in ability of bacterial strains to cause diseases of onion can be the result of genetic differences among strains of the same species. A bacterial cell reproduces by splitting into two perfect clones; this is often called vertical transfer of genes, analogous to genes inherited by children from their parents. However, bacteria also can trade snippets of genetic material (DNA) with each other, a process often called horizontal transfer of genes.
Usually, bacteria trade pieces of DNA in this manner between cells of the same or closely related species, but sometimes they can trade pieces of DNA with bacteria of other species. Even though DNA trading is relatively rare, it can have big consequences. For instance, bacterial gene trading is one of the ways that antibiotic resistance genes and genes for copper resistance spread among bacteria, undermining the effectiveness of antibiotics and bactericides. This is similar to the situation where some strains of Escherichia coli bacteria contain genes for toxins that give people food poisoning, whereas most strains of E. coli lack those genes and are harmless or even helpful members of the bacterial community in your digestive tract. Likewise, pathogenic and non-pathogenic bacteria can trade pieces of DNA when they are colonizing an onion together. This could turn the “saprophytes” (harmless bacteria) into pathogens of onion.
Developing DNA Diagnostic Tools
One of the overall goals of the Stop the Rot onion bacterial project is to develop diagnostic tools so that accurate diagnosis complements effective, pathogen-specific, targeted disease management strategies for various regions of onion production around the U.S. Currently, the primary method of diagnosing bacteria pathogenic to onion entails isolating bacteria from symptomatic tissue, and then characterizing the isolates and inoculating healthy plants to see if the isolates cause similar symptoms to what was observed on the original tissue. This can take weeks or even months to perform. DNA-based testing can be robust, rapid and relatively easy to standardize and can reduce the risks of “mistaken identity” with a non-pathogenic strain being diagnosed as the cause of the disease.
Sequencing the genome of a bacterial strain entails reading the individual letters (C, A, G and T) in the sequence of all the DNA in that bacterium. The sequenced genomes of pathogenic strains and non-pathogenic strains of the same species of bacterium then can be compared to identify specific genes found only in the pathogenic strains. For instance, we’ve learned that onion pathogenic strains of P. ananatis carry genes to synthesize the compound pantaphos, which is toxic to onion plants and allows the bacteria to cause onion leaf blight and bulb rot. The next step is to develop a DNA-based diagnostic tool that can be used to identify whether a bacterial strain contains the cluster of genes that produce pantaphos.
Progress
Developing DNA diagnostic tools requires a lot of testing to confirm the tool does not give false positive results (indicating a strain is a pathogen of onion when it is not) or false negative results (indicating a strain is not a pathogen when, in fact, it is). Since the DNA of every species of every genus of bacteria is different, the genes that confer pathogenicity in Pantoea agglomerans may not be the same as those in P. ananatis, Burkholderia species, and the many other bacteria that are pathogens of onion. Since there are numerous bacteria that can cause diseases of onion, developing DNA diagnostic tools for identifying specific bacteria that are pathogens of onion takes a lot of testing and time.
The Stop the Rot project has focused initially on developing diagnostic tools for strains of Pantoea that are pathogens of onion because this is one of the most common bacteria found in diseased onion crops and bulbs across the U.S. and many non-pathogenic strains of Pantoea also are found on onions. This research also has helped us identify strains of bacteria that carry genes for copper tolerance, i.e., genes that enable the bacteria to grow in the presence of copper. We are now testing whether the presence of bacteria with copper tolerance genes in onion crops might explain why applications of copper sprays in field trials across the U.S. typically have shown very poor control of bacterial diseases. We look forward to sharing these results in the near future.
Authors’ note: This work is supported by the Specialty Crops Research Initiative Award 2019-51181-30013 from the USDA National Institute of Food and Agriculture. Any opinions, findings, conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect the view of the U.S. Department of Agriculture.


